Developed MRI imaging Computer Vision algorithm using PyTorch, Matplotlib, and Numpy, to resample HBV and DCE T1-weighted images with respect to T2 images using rigid spatial transformation while utilizing real-world coordinates information.
Improved cancer classification accuracy by 11% compared to previous state of the art techniques, using in-house and global prostate MRI datasets; improved false-negative and true-positive detection
Code Sample
t2_mat = get_transformation_matrix('data/t2/')
new_t2_mat = np.array(t2_mat)
t2_img1_dcm = pydicom.dcmread('data/t2/IM-6392-0001.dcm')
t2_img1 = t2_img1_dcm.pixel_array
t2_img20_dcm = pydicom.dcmread('data/t2/IM-6392-0020.dcm')
t2_img20 = t2_img20_dcm.pixel_array
t2_h, t2_w = t2_img1_dcm.pixel_array.shape # 320*320
print(t2_h)
print(t2_w)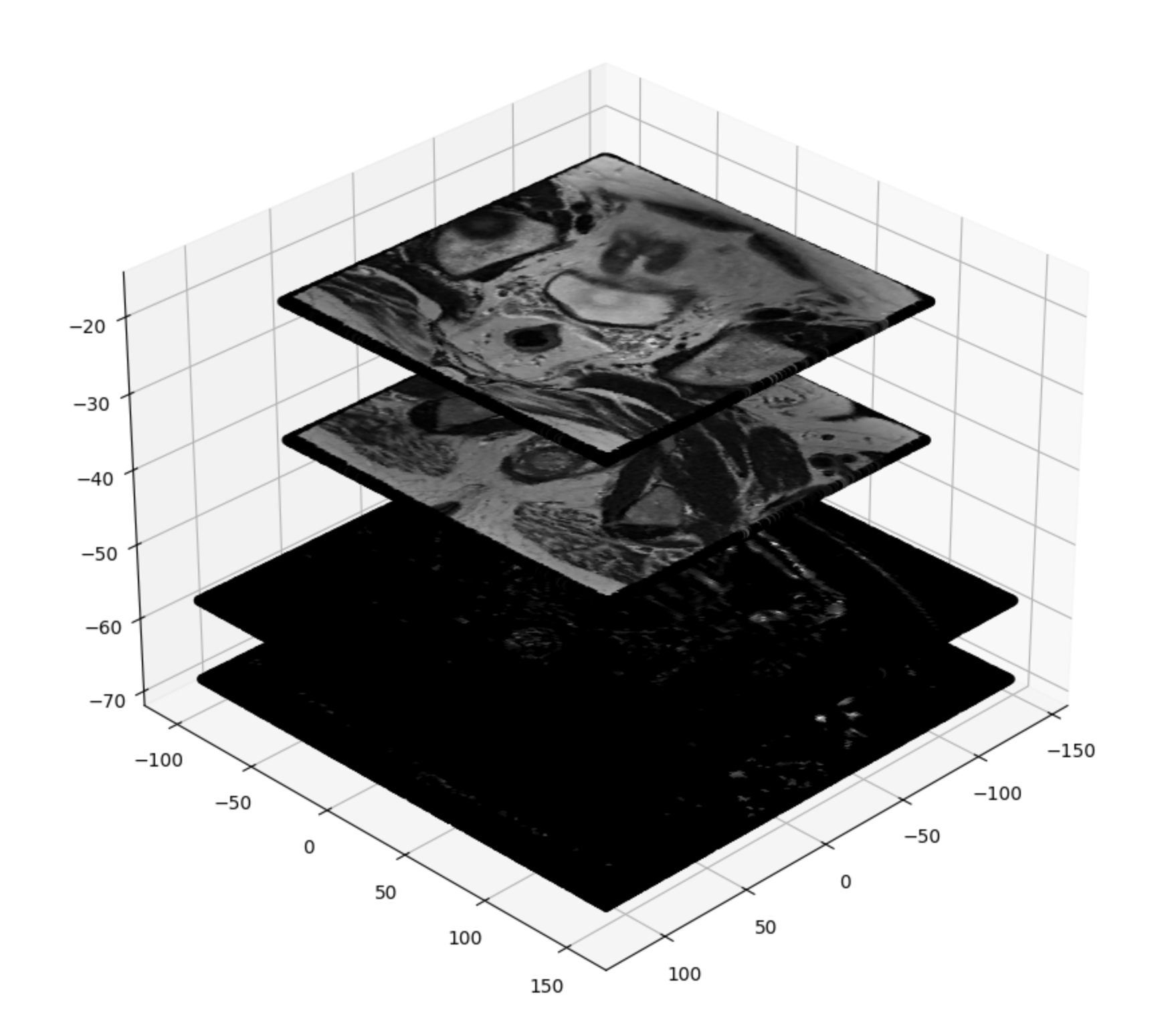
Credit
Parsa Hajipour
Kai Zhao